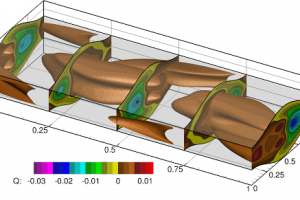
Nektar++ has been used to study various forms of hydrodynamic instabilities arising is a flow through a channel with corrugated walls. Stability analysis has been performed using direct numerical simulation and tracking growth, or attenuation of the unstable modes.
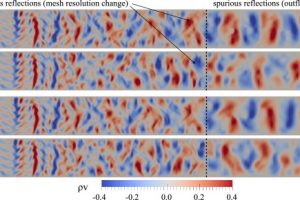
Nektar++ has been recently used to verify the dispersion/diffusion estimates of discontinuous spectral element methods for non-periodic flow problems. This work a key implications on the effective construction of LES tools for industrial problems.
Brief description of and Nektar++ installation instructions for Thomas, the UK National Tier 2 High Performance Computing Hub in Materials and Molecular Modelling.
We have recently performed some tests of the Nektar++ incompressible Navier-Stokes solver on a Formula 1 car geometry. The tests were run on Mira up to 131k cores and show excellent scaling of of the code for massively parallel simulations.
A Programme for the Nektar++ workshop is now available: Nektar++ Workshop 2017
Nektar++ has been used to investigate the incompressible flow around wings with spanwise waviness, using direct numerical simulations at different Reynolds numbers. The waviness was imposed by a coordinate transformation, using a novel technique presented in our JCP paper (1). This global mapping allowed us to consider a 2.5D problem, leading to significant advantages in terms […]
The Nektar++ Workshop 2017 will be held at Imperial College on 14th-15th June 2017. The 3rd annual Nektar++ Workshop will bring together developers and users of all experiences to hear about new and future developments in Nektar++ and the exciting science and engineering being undertaken with the code.
The latest version of Nektar++, v4.4.0, was released on the 9th March 2017. It can be downloaded from the downloads page.
DOI: 10.1016/j.jbiomech.2016.11.035
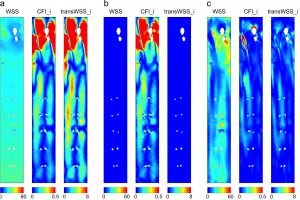
Here we use numerical methods to investigate the nature of the arterial flows captured by transWSS and the sensitivity of transWSS to inflow waveform and aortic geometry.
We encourage people who extend the code to help with their research to contribute those changes back into the main codebase for the benefit of other researchers. We have a number of tools and processes to help this including a code repository, a testing framework, code review and a continuous integration system.